|
|
BMe Research Grant |
|
George A. Olah Doctoral School of Chemistry and Chemical Technology
Faculty of Chemical Technology and Biotechnology, Department of Applied Biotechnology and Food Science
Supervisor: Dr. Vértessy G. Beáta
Gene expression of dUTPase isoforms
Introducing the research area
dUTPase is a key enzyme involved in the nucleotide metabolism and in the maintenance of genome integrity. Due to its physiological role, the enzyme is a promising target for cancer therapy. My research goal is to investigate the expression of dUTPase isoforms with RT-qPCR method in cancerous and normal human cells, mouse cells and mouse mammalian model. I determine the level of gene expression in different cell cycle phases and after serum starvation. My results may contribute to the increase of the effectiveness of chemotherapeutic treatments aiming the inhibition of dUTPase.
Brief introduction of the research place
I carry out my research at the Department of Applied Biotechnology and Food Science with the supervision of Prof. Dr. Beáta G. Vértessy. Our research group is involved in several areas of biology with a main focus on the enzyme dUTPase and on the presence and the role of uracil bases in DNA. Besides postdoctoral researchers and PhD students, many BSc and MSc students have also joined our group. We collaborate with several Hungarian and international partners.
History and context of the research
Both DNA and RNA are composed of nucleotides, however, DNA contains deoxyribose on the one hand, and uracil bases in RNA are replaced with thymine bases in DNA on the other. dUTPase is involved in the synthesis of thymine bases, and at the same time it is responsible for keeping uracil out of the genome1,2. On account of these functions, dUTPase is considered as a possible target for cancer therapy3–5, as dUTPase is most required by cells undergoing active proliferation6–10. Many publications corroborate that dUTPase protects cancerous cells from the chemotherapeutic agents 5-fluorouracil (5-FU) and 5-fluorodeoxyuridine (5-FdUr), which target the thymidylate biosynthesis. Moreover, increased expression of dUTPase may indicate a worse prognosis11–23. Considerable progress has been made in this field of cancer research: one small molecule inhibitor (TAS-114) has successfully passed clinical phase II trial24.
Two isoforms of dUTPase have been described in the literature, which catalyse the same reaction but their sequence is different3,25. One of them is the nuclear isoform, which locates in the nucleus, and is responsible for the synthesis of the DNA making up the vast majority of our genome. The other one is the mitochondrial isoform, which is essential for the synthesis of the short mitochondrial DNA encoding only 13 proteins, but which is continuously replicating in resting cells as well. Serum starvation forces cells to exit from the cell cycle and enter into a resting state. During this transition, the expression of the nuclear isoform decreases, while the expression of the mitochondrial isoform remains constant25.
The research goals, open questions
The expression of dUTPase was measured in many high-throughput studies, however, these studies cannot distinguish between the isoforms, and provide only the sum of the expression levels of the isoforms. With regard to the significance of the nuclear isoform, the isoform-specific examination is, however, essential to gain a valid and detailed insight into the respective physiological roles of the different isoforms and to design therapeutic applications based on this knowledge. Only two isoforms of human dUTPase have been described in the literature, however, recent high-throughput studies suggested the putative presence of two additional isoforms at both RNA and protein levels. The 3rd isoform does not contain any localisation signal, thus most probably remains in the cytosol that constitutes the bulk of the cells. The 4th isoform closely resembles the nuclear isoform, differing only in a few amino acids at the N-terminus but its expression is driven by an alternative promoter that may account for an altered regulation of this isoform. No data has been reported on the physiological or pathological roles of these two novel human dUTPase isoforms, however, the key biomedical significance of the dUTPase enzyme clearly necessitates further studies. In the current phase of my research, I am investigating the level of gene expression of the four isoforms of human dUTPase in various cancerous and normal human cell lines in order to understand the physiological role of the new isoforms.
Furthermore, I plan to analyse the expression levels of these isoforms during the cell cycle and during serum starvation to uncover the differences in the expression of dUTPase in different cell cycle phases and between actively proliferating and resting cells. These studies could shed light on the mechanisms underlying the sensitivity or resistance of these cells to chemotherapeutic agents.
Methods
To determine gene expression, I use a reverse transcription – quantitative polymerase chain reaction (RT-qPCR) method. This method mimics the natural DNA replication process, which is achieved in a series of cycles of temperature changes. The two DNA strands are separated at high temperature (95 °C), followed by a step at a lower temperature, in which oligonucleotides – the primers – marking the two ends of the DNA segment to be amplified are annealed to the single-stranded DNA templates. The heat stable DNA polymerase then completes the DNA segments with complementary bases, thus making two identical two-stranded segments. Then again the strands are separated at high temperature, thus doubling the amount of two-stranded DNA molecules in every cycle making an exponential amplification. To follow the qPCR reaction, a fluorescent dye is introduced, which binds to double-stranded DNA and its fluorescence is increased considerably as a consequence of its binding. The fluorescence is detected at the end of each cycle. To determine the relative amount of a specified DNA segment before the amplification in various samples, the number of cycles required for a threshold level of fluorescence is compared.
Gene expression measurements require the determination of messenger RNA molecules with a given sequence. Therefore, after the RNA isolation RNA molecules are transcribed to DNA with the use of a reverse transcriptase enzyme. The resulting complementary DNA (cDNA) can be used as a template in the qPCR reaction (Figure 1).
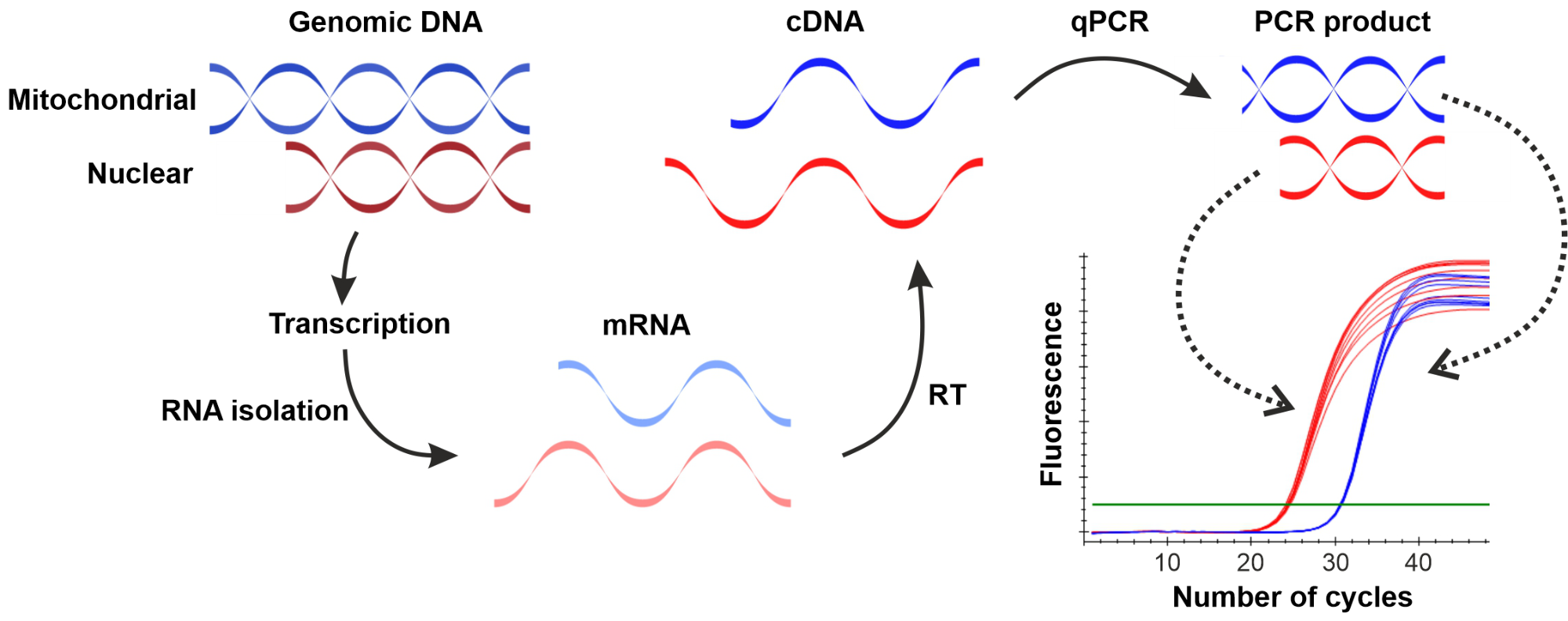
Figure 1: Schematic illustration of the RT-qPCR method used for the determination of gene expression
The development of a RT-qPCR method is a time and energy-consuming process, where multiple requirements have to be met. These are defined and specified in the “MIQE”-guidelines26. Since primers determine the DNA segments to be amplified, their design and selection is of utmost importance. One of the most critical controlled parameters is the annealing temperature, which greatly influences the specificity and efficiency of the reaction. The primer concentrations should also be optimised for maximum efficiency. The amount of RNA to be transcribed should also be determined to avoid inhibition. Amplification efficiency is a key performance indicator that measures the proportion of the DNA segments that doubles in each cycle. Investigating the specificity of the reaction is also crucial, which can be performed initially with agarose gel electrophoresis and product sequencing, and then routinely followed by melting curve analysis. Replicability and reproducibility can be determined by statistical analysis of variance.
To explore the cell cycle dependence of the dUTPase isoforms, the cells are treated with RO-3306 CDK1 inhibitor for 20 hours and synchronized in G2 phase. After the treatment and release, cells are collected every 2 hours and prepared for three experiments. The synchronous progression of cells through the cell cycle is followed by flow cytometry (FACS), gene expression levels are determined from isolated DNA, furthermore the protein amount of dUTPase is measured with Western-blot.
Results
In mice only two isoforms of dUTPase are present, whose expression was studied in different organs of male and female mice in my previous article10. The nuclear isoform is highly expressed in organs where active cell proliferation occurs (spleen, thymus, testicle, ovary), while the mitochondrial isoform is present in smaller amount but at a rather stable level in the organs examined (Figure 2). My results are in concordance with the research that has found a decrease of the expression of the nuclear isoform during serum starvation – where cell proliferation ceases – while the expression of the mitochondrial isoform remained constant as compared to freely proliferating cells25.
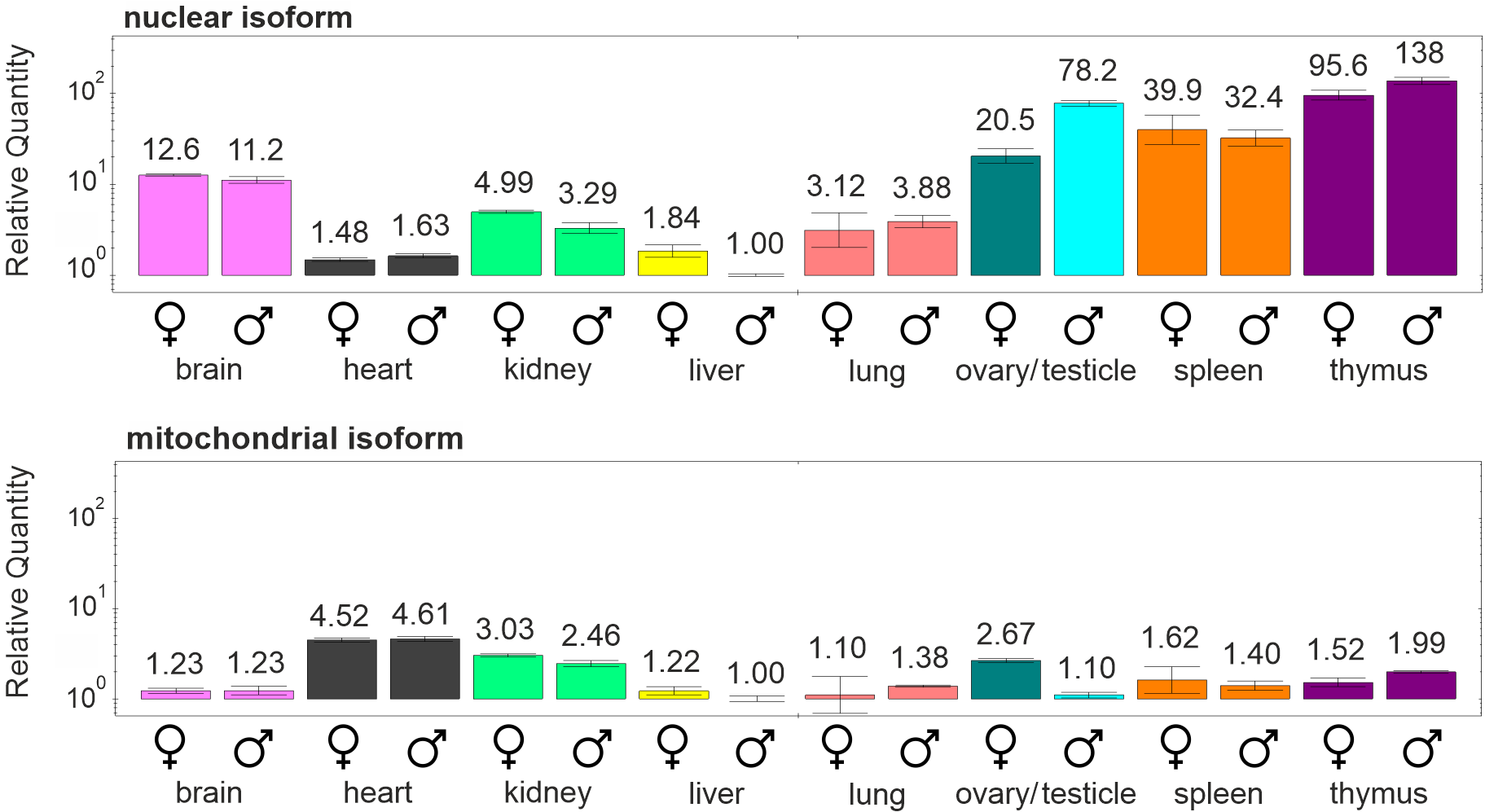
Figure 2: The expression levels of the nuclear and mitochondrial isoforms of dUTPase in different organs of male and female mice
I am currently developing a RT-qPCR method for the investigation of human dUTPase isoforms using my experience in method development. I confirmed the expression of all four isoforms; designed and selected the primers used for the amplification and determined the optimal annealing temperature and primer concentration for each target. In addition, I performed these steps for six reference genes as well. Reference genes are genes whose expression is relatively constant, independent of the characteristics of the cell lines. Expression levels of these genes can therefore constitute internal standard controls, thereby making it possible to compare the results obtained in different cell lines. The determination of PCR efficiencies is indispensable for the quantification of gene expression, and this parameter indicates the reliability of the reaction. Generally, efficiencies between 90% and 100% are acceptable. The efficiency values for the amplification of all isoforms and reference gene were measured to be between 96% and 100%. According to my preliminary results, in HCT-116 human colon cancer cell line it is the nuclear isoform that is expressed to the greatest extent, the expression of the mitochondrial isoform is around 4% of the nuclear isoform. The 3rd and 4th isoforms are expressed at approximately 2% and 0.4% of the nuclear isoform, respectively (Figure 3).
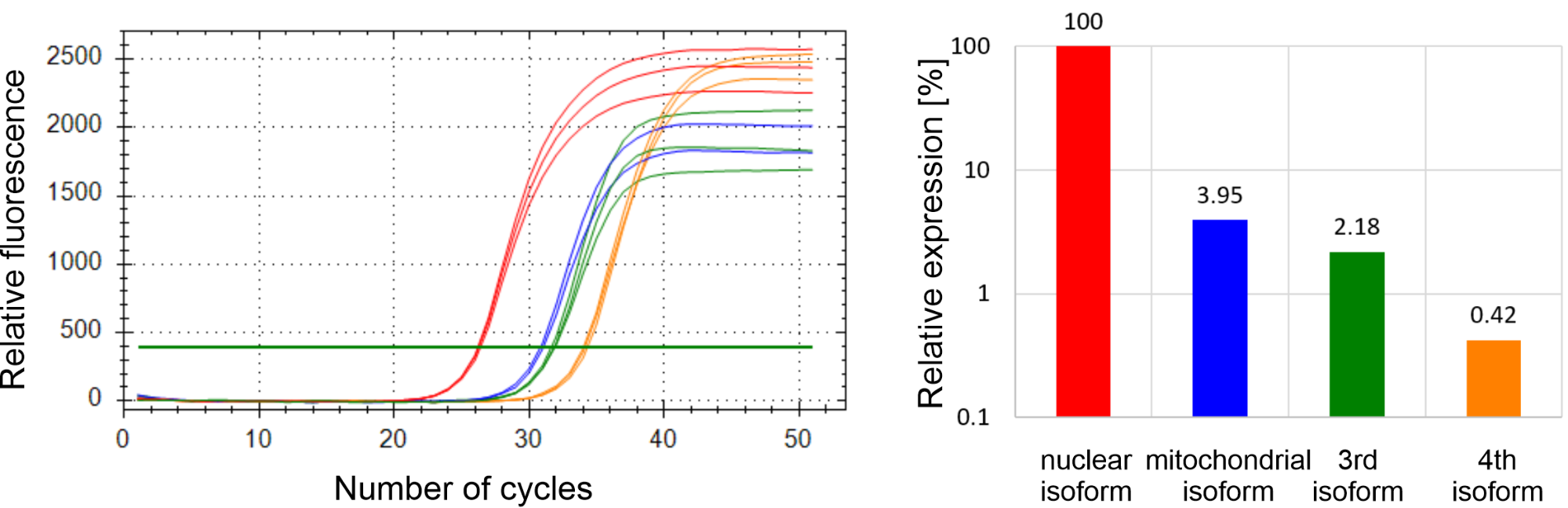
Figure 3: The expression levels of the four human dUTPase isoforms in HCT-116 cell line
A common assumption in the literature that the expression of the nuclear dUTPase isoform is coupled to the S phase of the cell cycle, i.e. to the DNA synthesis27. This assumption is based on a research article published in 1997 which investigated the expression changes of the two isoforms during serum starvation and subsequent release25. With serum starvation I acquired similar results, however, using the state of art method for synchronisation with the use of a selective CDK1 inhibitor, which stops the cells in the cell cycle but does not induce the cells to enter a resting state, I did not find any difference in the expression of the two isoforms in different cell cycle phases in mouse cells (Figure 4). I also investigated the expression at the protein level. I used actin as an internal standard and cyclin B1 as a cycle marker. Actin was present at a constant level, while the amount of cyclin B1 decreased gradually, then increased again, as expected, however, the level of dUTPase protein expression remained virtually constant (Figure 4). My experimental data therefore indicate that dUTPase expression is not S-phase-regulated, and it decreases only when cells enter the resting phase.
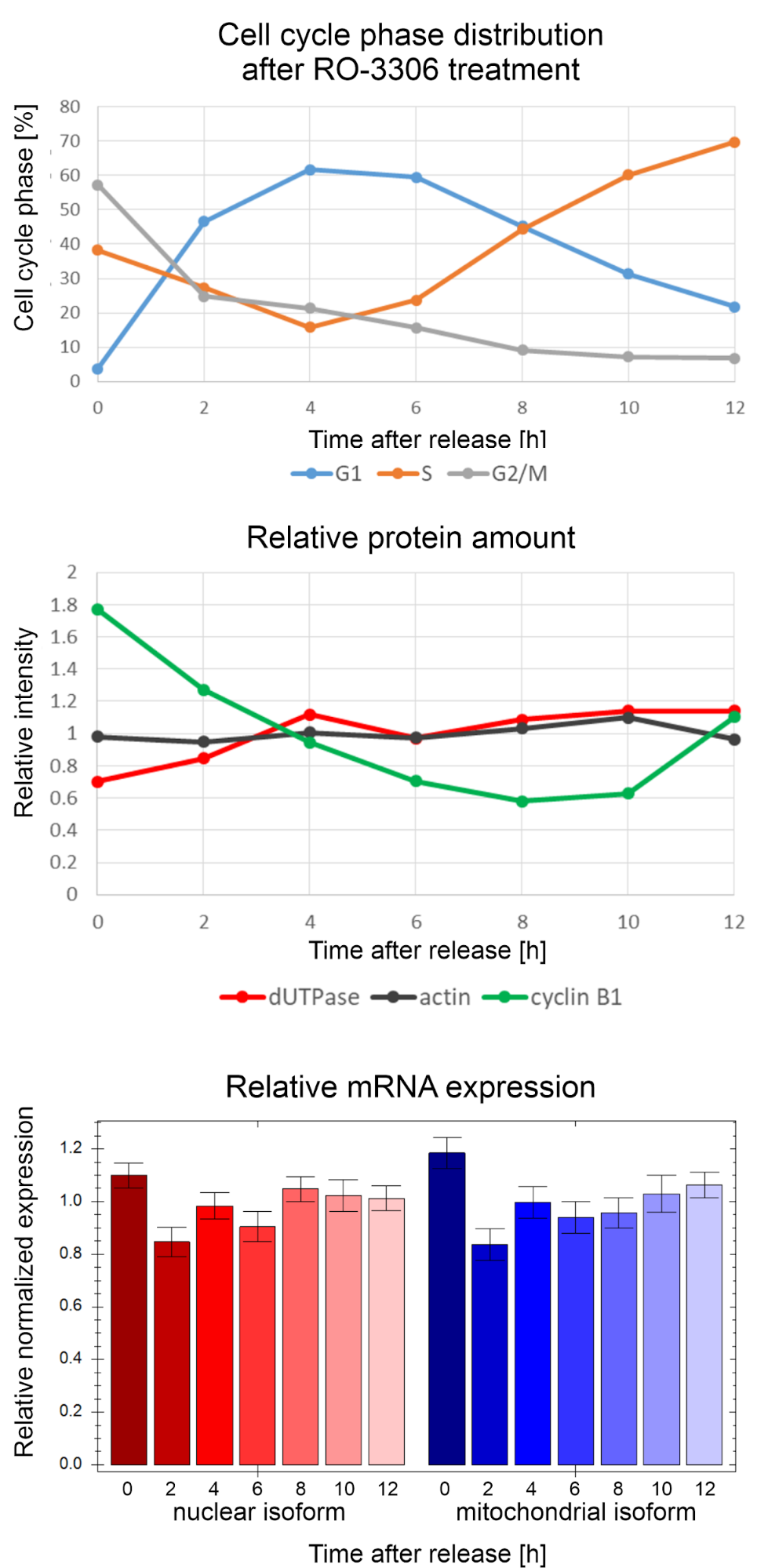
Figure 4: Gene expression of dUTPase on the protein and mRNA level in different phases of the cell cycle in mouse cells
Expected impact and further research
I measured the expression of all four isoforms of human dUTPase for the first time. With the developed RT-qPCR method, I plan to compare the expression levels of the isoforms in cell lines derived from various types of cancer and in normal cells. With the isoform-specific examination we can draw conclusions about the role of the previously unknown isoforms in cancerous cell lines. In mouse cells, the expression of the nuclear isoform is independent of the cell cycle phase, which is in contrast to the literature. The disproof of this assumption can contribute to a better understanding of the role of dUTPase in cellular metabolic processes, thus to the success of chemotherapeutic treatments aiming at dUTPase inhibition.
The newly developed method enables us to investigate the cell cycle dependence of all the four isoforms of dUTPase in human cells as well. Based on my results, I plan to publish two scientific articles in Q1-ranking journals
Publications, references, links
List of corresponding own publications
S1. Rácz, Gergely A.; Nagy, Nikolett; Gál, Zoltán; Pintér, Tímea; Hiripi, László; Vértessy, Beáta G. Evaluation of critical design parameters for RT-qPCR-based analysis of multiple dUTPase isoform genes in mice. FEBS Open Bio. Paper: 2211-5463.12654 (2019)
S2. Pálinkás, Hajnalka Laura; Rácz, Gergely Attila; Gál, Zoltán; Hoffmann, Orsolya Ivett; Tihanyi, Gergely; Róna, Gergely; Gócza, Elen; Hiripi, László; Vértessy, Beáta G. CRISPR/Cas9-Mediated Knock-Out of dUTPase in Mice Leads to Early Embryonic Lethality. Biomolecules 9:(4) Paper: 136 (2019)
Table of links
https://www.uniprot.org/uniprot/P33316
https://www.ncbi.nlm.nih.gov/gene/1854
https://www.uniprot.org/uniprot/A0A0C4DGL3
https://www.uniprot.org/uniprot/H0YNW5
https://en.wikipedia.org/wiki/Reverse_transcription_polymerase_chain_reaction
https://en.wikipedia.org/wiki/Polymerase_chain_reaction
https://en.wikipedia.org/wiki/Reverse_transcriptase
https://en.wikipedia.org/wiki/Agarose_gel_electrophoresis
https://en.wikipedia.org/wiki/Sanger_sequencing
https://en.wikipedia.org/wiki/Melting_curve_analysis
https://en.wikipedia.org/wiki/Analysis_of_variance
https://en.wikipedia.org/wiki/Cyclin-dependent_kinase_1
https://en.wikipedia.org/wiki/Flow_cytometry
https://en.wikipedia.org/wiki/Western_blot
List of references.
- Vértessy, B. G. & Tóth, J. Keeping Uracil Out of DNA: Physiological Role, Structure and Catalytic Mechanism of dUTPases. Acc. Chem. Res. 42, 97–106 (2009).
- Kerepesi, C. et al. Life without dUTPase. Front. Microbiol. 7, 1768 (2016).
- Ladner, R. D., McNulty, D. E., Carr, S. A., Roberts, G. D. & Caradonna, S. J. Characterization of Distinct Nuclear and Mitochondrial Forms of Human Deoxyuridine Triphosphate Nucleotidohydrolase. J. Biol. Chem. 271, 7745–7751 (1996).
- Mcintosh, E. M., Ager, D. D., Gadsden, M. H. & Haynest, R. H. Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc. Natl. Acad. Sci. 89, 8020–8024 (1992).
- Beck, W. R., Wright, G. E., Nusbaum, N. J., Chang, J. D. & Isselbacher, E. M. Enhancement of Methotrexate Cytotoxicity by Uracil Analogues that Inhibit Deoxyuridine Triphosphate Nucleotidohydrolase (dUTPase) Activity. in 97–104 (1986). doi:10.1007/978-1-4684-1248-2_16
- Pardo, E. G. & Gutiérrez, C. Cell cycle- and differentiation stage-dependent variation of dUTPase activity in higher plant cells. Exp. Cell Res. 186, 90–98 (1990).
- Pri-Hadash, A., Hareven, D. & Lifschitz, E. A meristem-related gene from tomato encodes a dUTPase: analysis of expression in vegetative and floral meristems. Plant Cell 4, 149–159 (1992).
- Strahler, J. R. et al. Maturation stage and proliferation-dependent expression of dUTPase in human T cells. Proc. Natl. Acad. Sci. 90, 4991–4995 (1993).
- Hokari, S., Sakagishi, Y. & Tsukada, K. Enhanced activity of deoxyuridine 5′-triphosphatase in regenerating rat liver. Biochem. Biophys. Res. Commun. 108, 95–101 (1982).
- Rácz, G. A. et al. Evaluation of critical design parameters for RT-qPCR-based analysis of multiple dUTPase isoform genes in mice. FEBS Open Bio 9, 1153–1170 (2019).
- Canman, C. E., Lawrence, T. S., Shewach, D. S., Tang, H. Y. & Maybaum, J. Resistance to fluorodeoxyuridine-induced DNA damage and cytotoxicity correlates with an elevation of deoxyuridine triphosphatase activity and failure to accumulate deoxyuridine triphosphate. Cancer Res. 53, 5219–24 (1993).
- Canman, C. E. et al. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 54, 2296–8 (1994).
- Fleischmann, J. et al. Expression of deoxyuridine triphosphatase (dUTPase) in colorectal tumours. Int. J. Cancer 84, 614–617 (1999).
- Romeike, B. F. M. et al. Immunohistochemical detection of dUTPase in intracranial tumors. Pathol. - Res. Pract. 201, 727–732 (2005).
- Wilson, P. M., LaBonte, M. J., Lenz, H.-J., Mack, P. C. & Ladner, R. D. Inhibition of dUTPase Induces Synthetic Lethality with Thymidylate Synthase-Targeted Therapies in Non-Small Cell Lung Cancer. Mol. Cancer Ther. 11, 616–628 (2012).
- Parsels, L. A. et al. Mechanism and pharmacological specificity of dUTPase-mediated protection from DNA damage and cytotoxicity in human tumor cells. Cancer Chemother. Pharmacol. 42, 357–362 (1998).
- Webley, S. D., Welsh, S. J., Jackman, A. L. & Aherne, G. W. The ability to accumulate deoxyuridine triphosphate and cellular response to thymidylate synthase (TS) inhibition. Br. J. Cancer 85, 446–452 (2001).
- Takatori, H. et al. dUTP pyrophosphatase expression correlates with a poor prognosis in hepatocellular carcinoma. Liver Int. 30, 438–446 (2009).
- Kawahara, A. et al. Higher expression of deoxyuridine triphosphatase (dUTPase) may predict the metastasis potential of colorectal cancer. J. Clin. Pathol. 62, 364–369 (2009).
- Nobili, S. et al. Identification of potential pharmacogenomic markers of clinical efficacy of 5-fluorouracil in colorectal cancer. Int. J. Cancer 128, 1935–1945 (2011).
- Webley, S. D., Hardcastle, A., Ladner, R. D., Jackman, A. L. & Aherne, G. W. Deoxyuridine triphosphatase (dUTPase) expression and sensitivity to the thymidylate synthase (TS) inhibitor ZD9331. Br. J. Cancer 83, 792–799 (2000).
- Ladner, R. D. et al. dUTP nucleotidohydrolase isoform expression in normal and neoplastic tissues: association with survival and response to 5-fluorouracil in colorectal cancer. Cancer Res. 60, 3493–503 (2000).
- Wilson, P. M. et al. Novel opportunities for thymidylate metabolism as a therapeutic target. Mol. Cancer Ther. 7, 3029–3037 (2008).
- Yamamoto, N. et al. A randomized, phase 2 study of deoxyuridine triphosphatase inhibitor, TAS-114, in combination with S-1 versus S-1 alone in patients with advanced non-small-cell lung cancer. Invest. New Drugs (2020). doi:10.1007/s10637-020-00930-5
- Ladner, R. D. & Caradonna, S. J. The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J. Biol. Chem. 272, 19072–19080 (1997).
- Bustin, S. A. et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 55, 611–622 (2009).
Wilson, P. M., Fazzone, W., LaBonte, M. J., Lenz, H. & Ladner, R. D. Regulation of human dUTPase gene expression and p53-mediated transcriptional repression in response to oxaliplatin-induced DNA damage. Nucleic Acids Res. 37, 78–95 (2009).
